In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.
Post-translational modifications can occur on the amino acid side chains or at the protein's C- or N- termini.[1] They can expand the chemical set of the 22 amino acids by changing an existing functional group or adding a new one such as phosphate. Phosphorylation is highly effective for controlling the enzyme activity and is the most common change after translation. [2] Many eukaryotic and prokaryotic proteins also have carbohydrate molecules attached to them in a process called glycosylation, which can promote protein folding and improve stability as well as serving regulatory functions. Attachment of lipid molecules, known as lipidation, often targets a protein or part of a protein attached to the cell membrane.
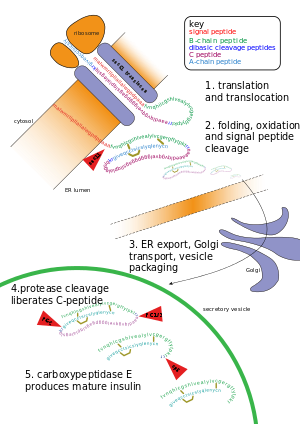
Other forms of post-translational modification consist of cleaving peptide bonds, as in processing a propeptide to a mature form or removing the initiator methionine residue. The formation of disulfide bonds from cysteine residues may also be referred to as a post-translational modification.[3] For instance, the peptide hormone insulin is cut twice after disulfide bonds are formed, and a propeptide is removed from the middle of the chain; the resulting protein consists of two polypeptide chains connected by disulfide bonds.
Some types of post-translational modification are consequences of oxidative stress. Carbonylation is one example that targets the modified protein for degradation and can result in the formation of protein aggregates.[4][5] Specific amino acid modifications can be used as biomarkers indicating oxidative damage.[6]
Sites that often undergo post-translational modification are those that have a functional group that can serve as a nucleophile in the reaction: the hydroxyl groups of serine, threonine, and tyrosine; the amine forms of lysine, arginine, and histidine; the thiolate anion of cysteine; the carboxylates of aspartate and glutamate; and the N- and C-termini. In addition, although the amide of asparagine is a weak nucleophile, it can serve as an attachment point for glycans. Rarer modifications can occur at oxidized methionines and at some methylene groups in side chains.[7]
Post-translational modification of proteins can be experimentally detected by a variety of techniques, including mass spectrometry, Eastern blotting, and Western blotting. Additional methods are provided in the #External links section.
PTMs involving addition of functional groups edit
Addition by an enzyme in vivo edit
Hydrophobic groups for membrane localization edit
- myristoylation (a type of acylation), attachment of myristate, a C14 saturated acid
- palmitoylation (a type of acylation), attachment of palmitate, a C16 saturated acid
- isoprenylation or prenylation, the addition of an isoprenoid group (e.g. farnesol and geranylgeraniol)
- glypiation, glycosylphosphatidylinositol (GPI) anchor formation via an amide bond to C-terminal tail
Cofactors for enhanced enzymatic activity edit
- lipoylation (a type of acylation), attachment of a lipoate (C8) functional group
- flavin moiety (FMN or FAD) may be covalently attached
- heme C attachment via thioether bonds with cysteines
- phosphopantetheinylation, the addition of a 4'-phosphopantetheinyl moiety from coenzyme A, as in fatty acid, polyketide, non-ribosomal peptide and leucine biosynthesis
- retinylidene Schiff base formation
Modifications of translation factors edit
- diphthamide formation (on a histidine found in eEF2)
- ethanolamine phosphoglycerol attachment (on glutamate found in eEF1α)[8]
- hypusine formation (on conserved lysine of eIF5A (eukaryotic) and aIF5A (archaeal))
- beta-Lysine addition on a conserved lysine of the elongation factor P (EFP) in most bacteria.[9] EFP is a homolog to eIF5A (eukaryotic) and aIF5A (archaeal) (see above).
Smaller chemical groups edit
- acylation, e.g. O-acylation (esters), N-acylation (amides), S-acylation (thioesters)
- acetylation, the addition of an acetyl group, either at the N-terminus of the protein or at lysine residues.[10] The reverse is called deacetylation.
- formylation
- alkylation, the addition of an alkyl group, e.g. methyl, ethyl
- methylation the addition of a methyl group, usually at lysine or arginine residues. The reverse is called demethylation.
- amidation at C-terminus. Formed by oxidative dissociation of a C-terminal Gly residue.[11]
- amide bond formation
- amino acid addition
- arginylation, a tRNA-mediation addition
- polyglutamylation, covalent linkage of glutamic acid residues to the N-terminus of tubulin and some other proteins.[12] (See tubulin polyglutamylase)
- polyglycylation, covalent linkage of one to more than 40 glycine residues to the tubulin C-terminal tail
- amino acid addition
- butyrylation
- gamma-carboxylation dependent on Vitamin K[13]
- glycosylation, the addition of a glycosyl group to either arginine, asparagine, cysteine, hydroxylysine, serine, threonine, tyrosine, or tryptophan resulting in a glycoprotein. Distinct from glycation, which is regarded as a nonenzymatic attachment of sugars.
- O-GlcNAc, addition of N-acetylglucosamine to serine or threonine residues in a β-glycosidic linkage
- polysialylation, addition of polysialic acid, PSA, to NCAM
- malonylation
- hydroxylation: addition of an oxygen atom to the side-chain of a Pro or Lys residue
- iodination: addition of an iodine atom to the aromatic ring of a tyrosine residue (e.g. in thyroglobulin)
- nucleotide addition such as ADP-ribosylation
- phosphate ester (O-linked) or phosphoramidate (N-linked) formation
- phosphorylation, the addition of a phosphate group, usually to serine, threonine, and tyrosine (O-linked), or histidine (N-linked)
- adenylylation, the addition of an adenylyl moiety, usually to tyrosine (O-linked), or histidine and lysine (N-linked)
- uridylylation, the addition of an uridylyl-group (i.e. uridine monophosphate, UMP), usually to tyrosine
- propionylation
- pyroglutamate formation
- S-glutathionylation
- S-nitrosylation
- S-sulfenylation (aka S-sulphenylation), reversible covalent addition of one oxygen atom to the thiol group of a cysteine residue[14]
- S-sulfinylation, normally irreversible covalent addition of two oxygen atoms to the thiol group of a cysteine residue[14]
- S-sulfonylation, normally irreversible covalent addition of three oxygen atoms to the thiol group of a cysteine residue, resulting in the formation of a cysteic acid residue[14]
- succinylation addition of a succinyl group to lysine
- sulfation, the addition of a sulfate group to a tyrosine.
Non-enzymatic modifications in vivo edit
Examples of non-enzymatic PTMs are glycation, glycoxidation, nitrosylation, oxidation, succination, and lipoxidation.[15]
- glycation, the addition of a sugar molecule to a protein without the controlling action of an enzyme.
- carbamylation the addition of Isocyanic acid to a protein's N-terminus or the side-chain of Lys.[16]
- carbonylation the addition of carbon monoxide to other organic/inorganic compounds.
- spontaneous isopeptide bond formation, as found in many surface proteins of Gram-positive bacteria.[17]
Non-enzymatic additions in vitro edit
- biotinylation: covalent attachment of a biotin moiety using a biotinylation reagent, typically for the purpose of labeling a protein.
- carbamylation: the addition of Isocyanic acid to a protein's N-terminus or the side-chain of Lys or Cys residues, typically resulting from exposure to urea solutions.[18]
- oxidation: addition of one or more Oxygen atoms to a susceptible side-chain, principally of Met, Trp, His or Cys residues. Formation of disulfide bonds between Cys residues.
- pegylation: covalent attachment of polyethylene glycol (PEG) using a pegylation reagent, typically to the N-terminus or the side-chains of Lys residues. Pegylation is used to improve the efficacy of protein pharmaceuticals.
Conjugation with other proteins or peptides edit
- ubiquitination, the covalent linkage to the protein ubiquitin.
- SUMOylation, the covalent linkage to the SUMO protein (Small Ubiquitin-related MOdifier)[19]
- neddylation, the covalent linkage to the Nedd protein
- ISGylation, the covalent linkage to the ISG15 protein (Interferon-Stimulated Gene 15)[20]
- pupylation, the covalent linkage to the prokaryotic ubiquitin-like protein
Chemical modification of amino acids edit
- citrullination, or deimination, the conversion of arginine to citrulline[21]
- deamidation, the conversion of glutamine to glutamic acid or asparagine to aspartic acid
- eliminylation, the conversion to an alkene by beta-elimination of phosphothreonine and phosphoserine, or dehydration of threonine and serine[22]
Structural changes edit
- disulfide bridges, the covalent linkage of two cysteine amino acids
- lysine-cysteine bridges, the covalent linkage of 1 lysine and 1 or 2 cystine residues via an oxygen atom (NOS and SONOS bridges)[23]
- proteolytic cleavage, cleavage of a protein at a peptide bond
- isoaspartate formation, via the cyclisation of asparagine or aspartic acid amino-acid residues
- racemization
- of serine by protein-serine epimerase
- of alanine in dermorphin, a frog opioid peptide
- of methionine in deltorphin, also a frog opioid peptide
- protein splicing, self-catalytic removal of inteins analogous to mRNA processing
Statistics edit
Common PTMs by frequency edit
In 2011, statistics of each post-translational modification experimentally and putatively detected have been compiled using proteome-wide information from the Swiss-Prot database.[24] The 10 most common experimentally found modifications were as follows:[25]
| Frequency | Modification |
|---|---|
| 58383 | Phosphorylation |
| 6751 | Acetylation |
| 5526 | N-linked glycosylation |
| 2844 | Amidation |
| 1619 | Hydroxylation |
| 1523 | Methylation |
| 1133 | O-linked glycosylation |
| 878 | Ubiquitylation |
| 826 | Pyrrolidone carboxylic acid |
| 504 | Sulfation |
Common PTMs by residue edit
Some common post-translational modifications to specific amino-acid residues are shown below. Modifications occur on the side-chain unless indicated otherwise.
Databases and tools edit

Protein sequences contain sequence motifs that are recognized by modifying enzymes, and which can be documented or predicted in PTM databases. With the large number of different modifications being discovered, there is a need to document this sort of information in databases. PTM information can be collected through experimental means or predicted from high-quality, manually curated data. Numerous databases have been created, often with a focus on certain taxonomic groups (e.g. human proteins) or other features.
List of resources edit
- PhosphoSitePlus[27] – A database of comprehensive information and tools for the study of mammalian protein post-translational modification
- ProteomeScout[28] – A database of proteins and post-translational modifications experimentally
- Human Protein Reference Database[28] – A database for different modifications and understand different proteins, their class, and function/process related to disease causing proteins
- PROSITE[29] – A database of Consensus patterns for many types of PTM's including sites
- RESID[30] – A database consisting of a collection of annotations and structures for PTMs.
- iPTMnet [31]– A database that integrates PTM information from several knowledgbases and text mining results.
- dbPTM[26] – A database that shows different PTM's and information regarding their chemical components/structures and a frequency for amino acid modified site
- Uniprot has PTM information although that may be less comprehensive than in more specialized databases.

Effect of PTMs on protein function and physiological processes.[32] - The O-GlcNAc Database[33][34] - A curated database for protein O-GlcNAcylation and referencing more than 14 000 protein entries and 10 000 O-GlcNAc sites.
Tools edit
List of software for visualization of proteins and their PTMs
Case examples edit
This section needs additional citations for verification. (January 2016) |
- Cleavage and formation of disulfide bridges during the production of insulin
- PTM of histones as regulation of transcription: RNA polymerase control by chromatin structure
- PTM of RNA polymerase II as regulation of transcription
- Cleavage of polypeptide chains as crucial for lectin specificity[38]
See also edit
References edit
External links edit
(Wayback Machine copy)
- List of posttranslational modifications in ExPASy
- Browse SCOP domains by PTM — from the dcGO database
- Statistics of each post-translational modification from the Swiss-Prot database
(Wayback Machine copy)
- AutoMotif Server - A Computational Protocol for Identification of Post-Translational Modifications in Protein Sequences
- Functional analyses for site-specific phosphorylation of a target protein in cells
- Detection of Post-Translational Modifications after high-accuracy MSMS
- Overview and description of commonly used post-translational modification detection techniques

